The history of single cell sequencing
Single cell sequencing (SCS) technology refers to the sequencing of the genetic information carried by cells at the level of a single cell, aiming to obtain gene sequences, transcripts, proteins and epigenetic information of a certain cell type and conduct the integrated analysis. It has been widely used in new species identification, pathogen screening, pathogen evolution, developmental biology, neuroscience, tumor heterogeneity research and circulating tumor cells [1-2]. In 2013, single-cell sequencing technology was named Technology of the Year by Nature Methods. In the same year, Science ranked the single-cell sequence at the top of the six most noteworthy areas of the year.
Since the publication of the first single-cell transcriptome research article in 2009, single-cell sequencing has undergone more than ten years of development. With the continuous improvement of a variety of technologies and the emergence of new technologies, for example, the introduction of microfluidics, random capture methods, and in-situ barcodes has promoted the cost reduction of reagents and consumables, and the scale of sequencing has been also increased from ~100 cells to hundreds of thousands or even millions of cells.
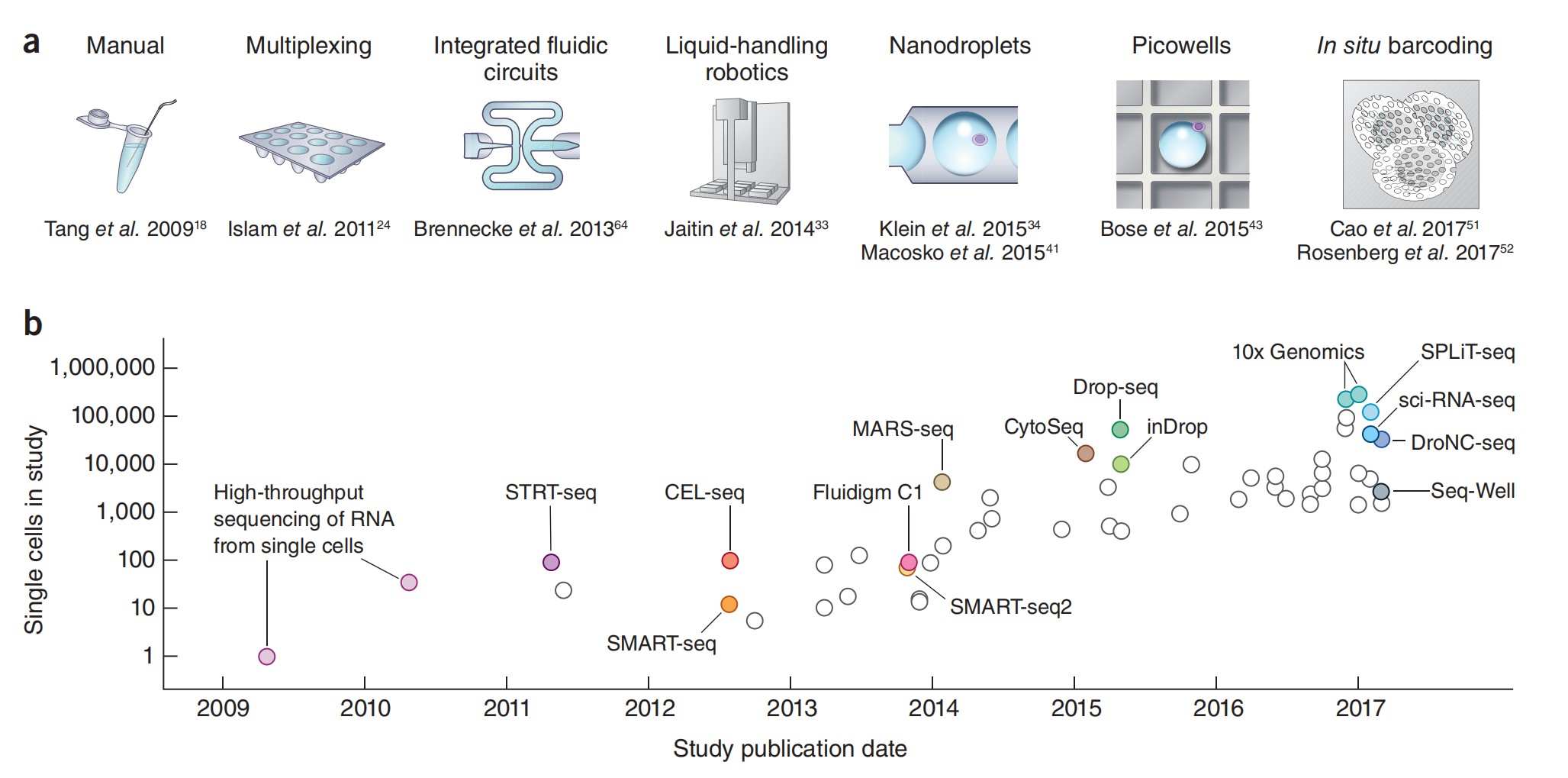
Figure 1:Scaling of scRNA-seq experiments.。(a) Key technologies that have allowed jumps in experimental scale; (b) Cell numbers reported in representative publications by publication date[3].
How to achieve cell-level sequencing?
There are about 40-60 trillion cells in the human body, and the average cell diameter is between 5 and 200 microns. To sequence a single cell, the first strategy that comes to mind is to isolate the single cell, build a sequencing library independently, and finally perform the sequencing. There are a variety of methods that can be used to accurately manipulate/separate and analyze single cells, including flow cytometry, microfluidic systems, and various methods for separating and detecting single cells in micromodule systems.
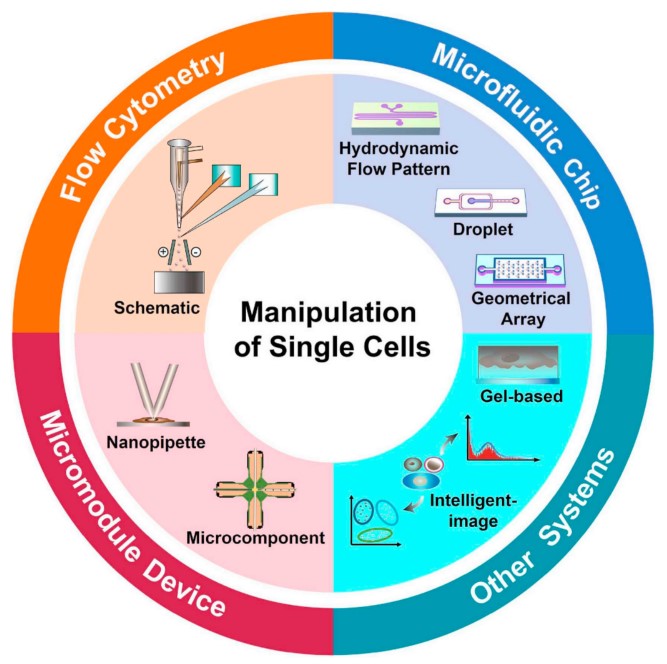
Figure 2: Classification of various strategies for isolation and manipulation of single cells[4].
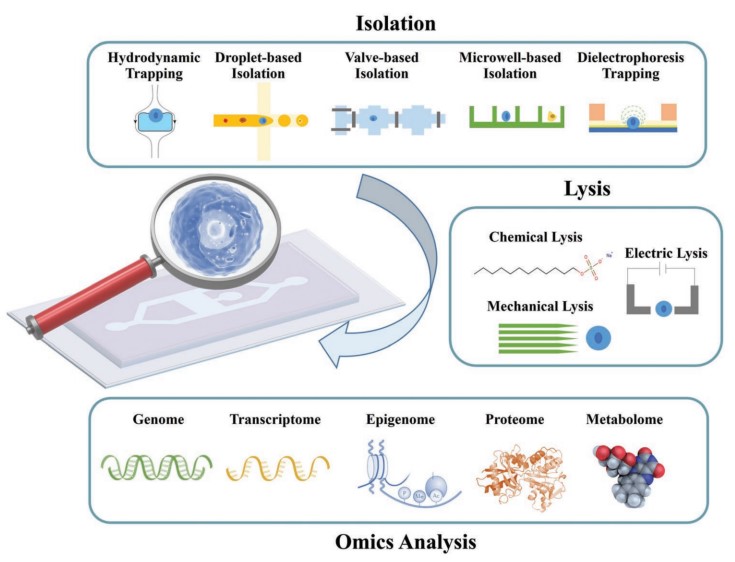
Figure 3: Schematic illustration of the microfluidic single-cell omics analysis[5].
However, this strategy saves cell samples, but the high cost limits the increase in throughput and its wide application. In recent years, commercial single-cell sequencing platforms, such as BD Rhapsody and 10X Genomics, have introduced barcode-based single-cell identification. Add a unique DNA sequence barcode to each cell, and nucleic acid molecules carrying the same DNA sequence can be considered to come from the same cell. Using this strategy greatly simplifies the operation process of the sequencing library construction, and the information of hundreds of thousands of cells can be obtained in the same library.
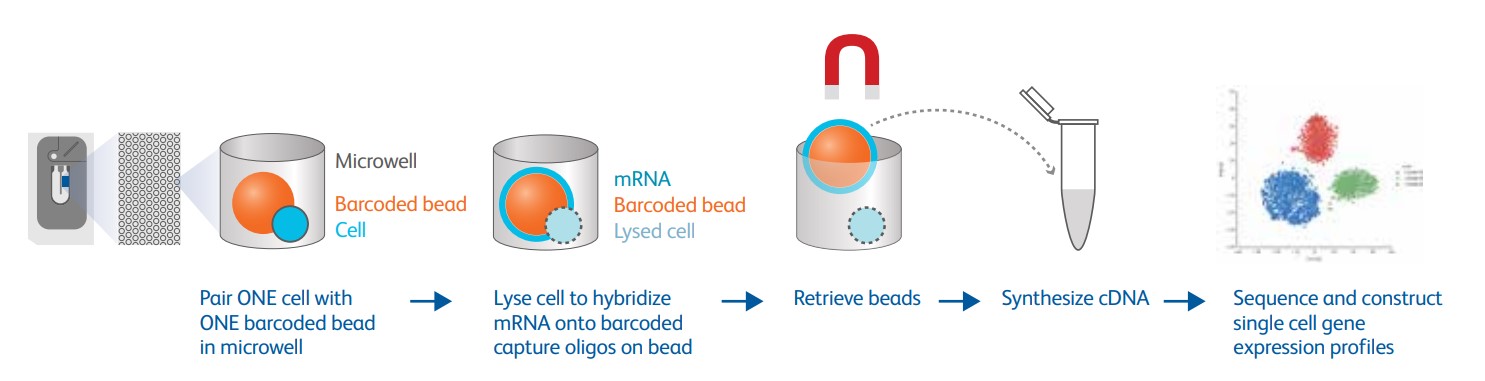
Figure 4: The workflow of BD Rhapsody: one microwell=one cell=one magnetic bead=one RNA-Seq (from BD official website)
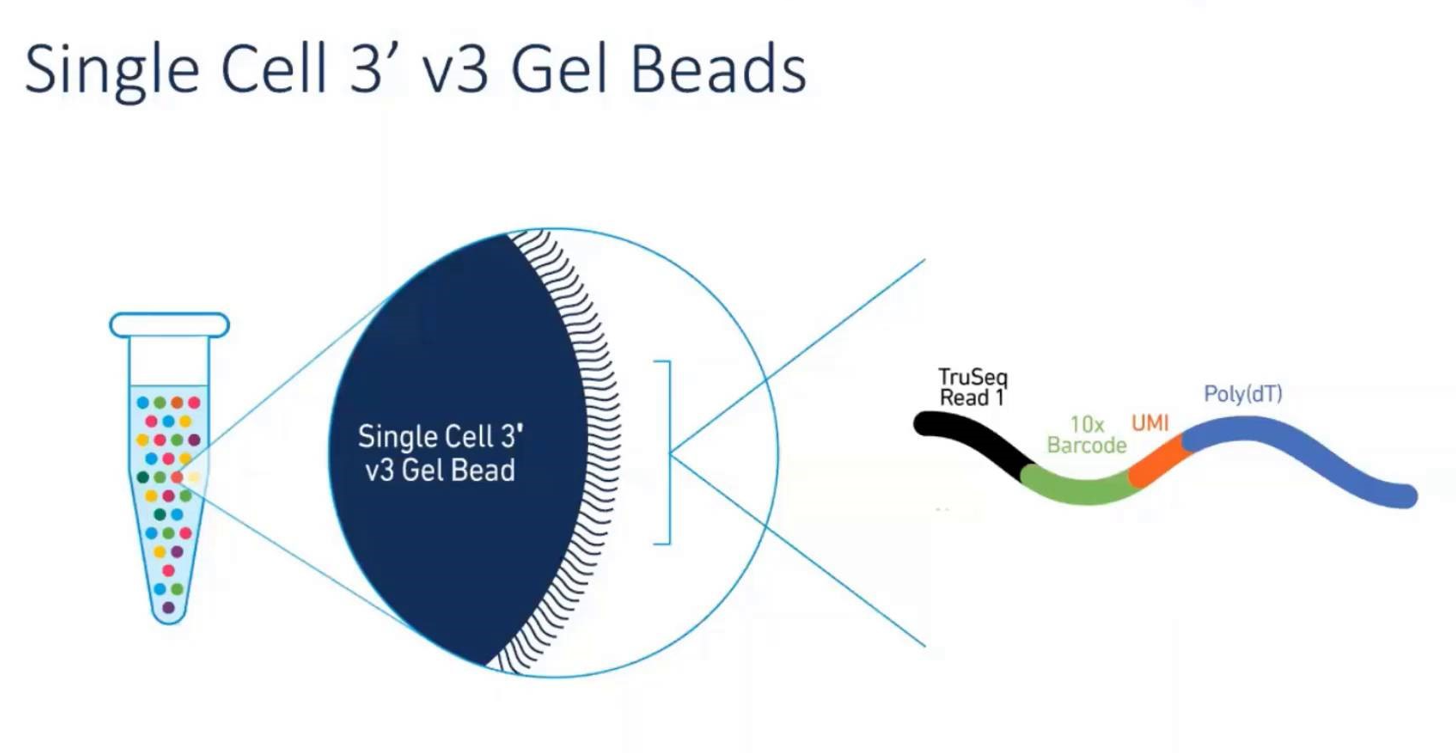
Figure 5: The workflow of 10X Genomics: one oil droplet=one cell=one gel bead=one RNA-Seq (from 10X Genomics official website)

